Rabix Executor Quickstart
Overview
Rabix is an open-source development project for creating and running computational workflows. It was founded by Seven Bridges with the goal of empowering researchers and developers to easily run reproducible analyses at scale. The Rabix Executor is used to run workflows described using the Common Workflow Language (CWL). It is currently runnable from the command line and is suitable for local testing and development.
Prerequisites
- Docker
- Java Development Kit or Java Runtime Environment (version 8)
Downloading and installing Rabix Executor
- Open the terminal on your computer.
- Enter the following command:
wget https://github.com/rabix/bunny/releases/download/v1.0.1/rabix-1.0.1.tar.gz && tar -zxvf rabix-1.0.1.tar.gzIf you are using macOS, you can install
wgetthrough a package manager such as Homebrew. - Once the download and unpacking are done, use the
cdcommand to navigate to the folder containing the downloaded package:cd rabix-cli-1.0.1You are now ready to start using the Rabix Executor.
Executing the example
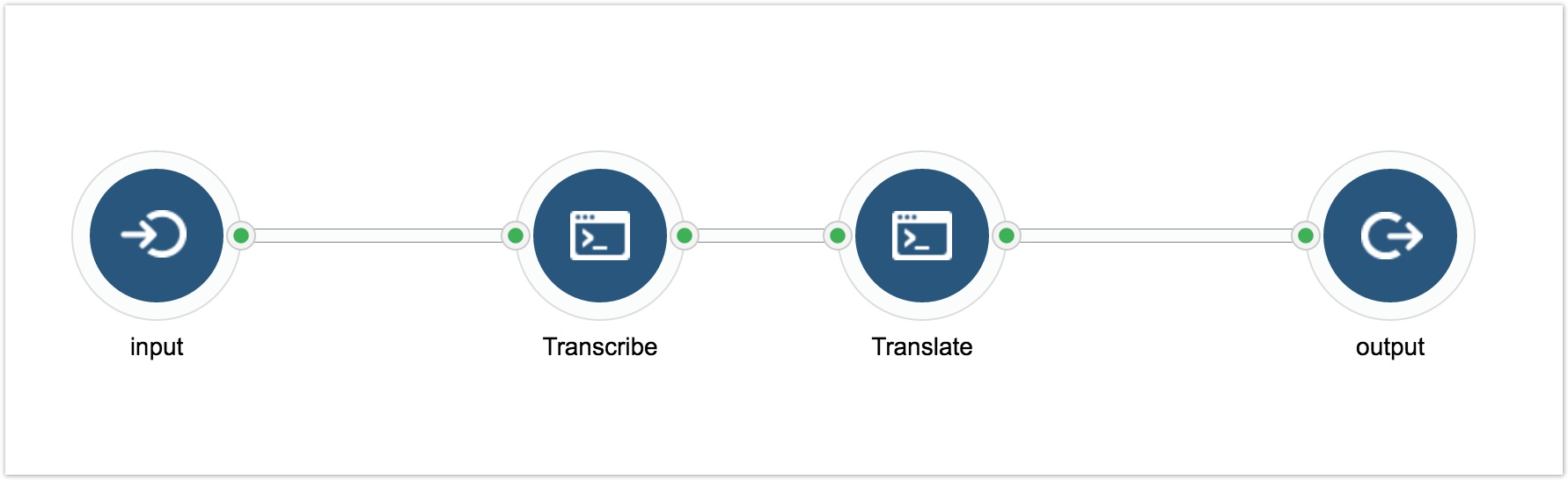
The downloaded .tar.gz package also includes a folder with sample CWL files that you can run to see a fully functional example of local execution. The example consists of two CWL tools (Transcribe, Translate) and one workflow (dna2protein). Transcribe takes a TXT file containing a DNA Sequence as an input and produces a TXT file with an mRNA Sequence. Translate takes an mRNA Sequence, identifies the first ORF, and produces a TXT file with a peptide sequence as an output. The dna2protein workflow includes both of those tools and they are set up as follows:
-
In this example we will execute the dna2protein workflow:
- In the folder where the Rabix Executor package has been unpacked execute the following command:
./rabix examples/dna2protein/dna2protein.cwl.json examples/dna2protein/inputs.jsonThis will execute the dna2protein workflow described above. Once the execution has been completed, the output is displayed in the terminal. You should see something similar to:
{ "output_protein" : { "checksum" : "sha1$55adf0ec2ecc6aee57a774d48216ac5a97d6e5ba", "class" : "File", "location" : "/Users/rfranklin/Rabix/rabix-cli-1.0.1/examples/dna2protein/dna2protein.cwl-20170404145015326/root/Translate/protein.txt", "name" : "protein.txt", "path" : "/Users/rfranklin/Rabix/rabix-cli-1.0.1/examples/dna2protein/dna2protein.cwl-20170404145015326/root/Translate/protein.txt", "secondaryFiles" : [ ], "size" : 9 } }The path to your output file is provided in the “path” field.
- Copy the path and execute the following command in the terminal, making sure to replace the path below with the one you copied in the previous step.
less /Users/rfranklin/Rabix/rabix-cli-1.0.1/examples/dna2protein/dna2protein.cwl-20170404145015326/root/Translate/protein.txtYou can see the content of the output file produced by this sample workflow. Press Q on the keyboard to return to your terminal screen.